Marker based heritability calculation
library(tidyverse)
library(GS)
library(sommer)
data(DT_cpdata)
geno <- GT_cpdata
samp <- rownames(GT_cpdata)
phen <- DT_cpdata tmp <- herMarker(geno, samp, phen, method = c("RKHS","sommer"), traits=c("color","Yield"))
# []==============================================================[]
# []======== Marker based heritability calculation ===============[]
# []================= BGLR - sommer package =====================[]
# []======= Last update: 2020-03-22 Johan Aparicio ==============[]
# []==============================================================[]
# color == 362
# Yield == 362
#
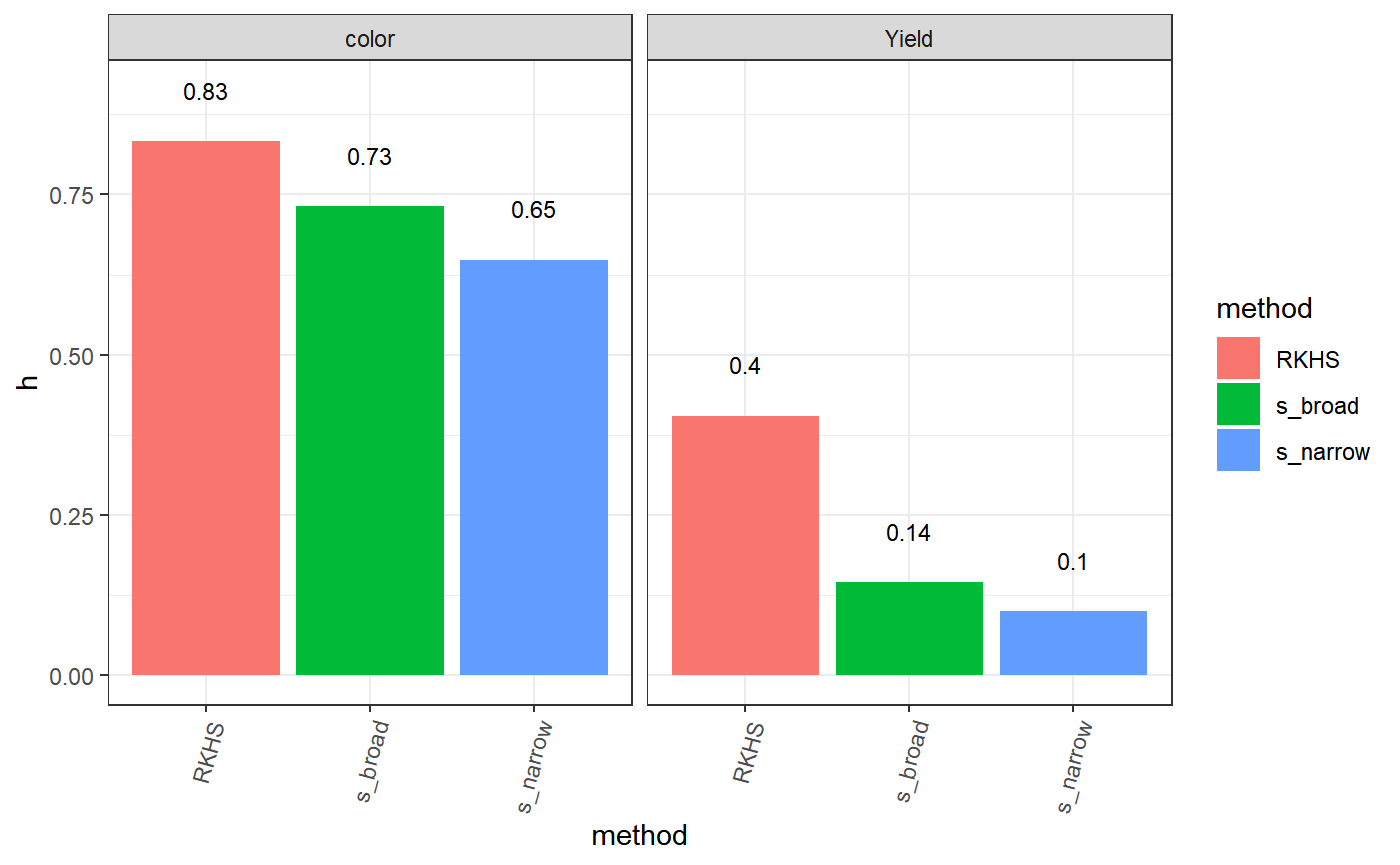
# trait method h corr finishedAt
# color RKHS 0.833 0.941 2020-04-02 21:38:09
# color s_narrow 0.648 0.87 2020-04-02 21:38:10
# color s_broad 0.732 0.861 2020-04-02 21:38:10
# Yield RKHS 0.405 0.71 2020-04-02 21:38:17
# Yield s_narrow 0.1 0.547 2020-04-02 21:38:18
# Yield s_broad 0.145 0.604 2020-04-02 21:38:18
#
# []============================ End =============================[] tmp$data[,-c(4,5)] %>% spread(method,h)
# trait RKHS s_broad s_narrow
# 1 color 0.833 0.732 0.648
# 2 Yield 0.405 0.145 0.100 tmp$data %>%
ggplot(aes(x=method, y=h,fill=method, label=round(h,2)))+
geom_bar(stat = "identity", position = "dodge" )+
theme_bw()+
theme(axis.text.x = element_text(hjust = 1,angle = 75))+
geom_text(aes(method), size=3,nudge_y = 0.08)+
facet_wrap(~trait,ncol = 2,scales = "free_x")