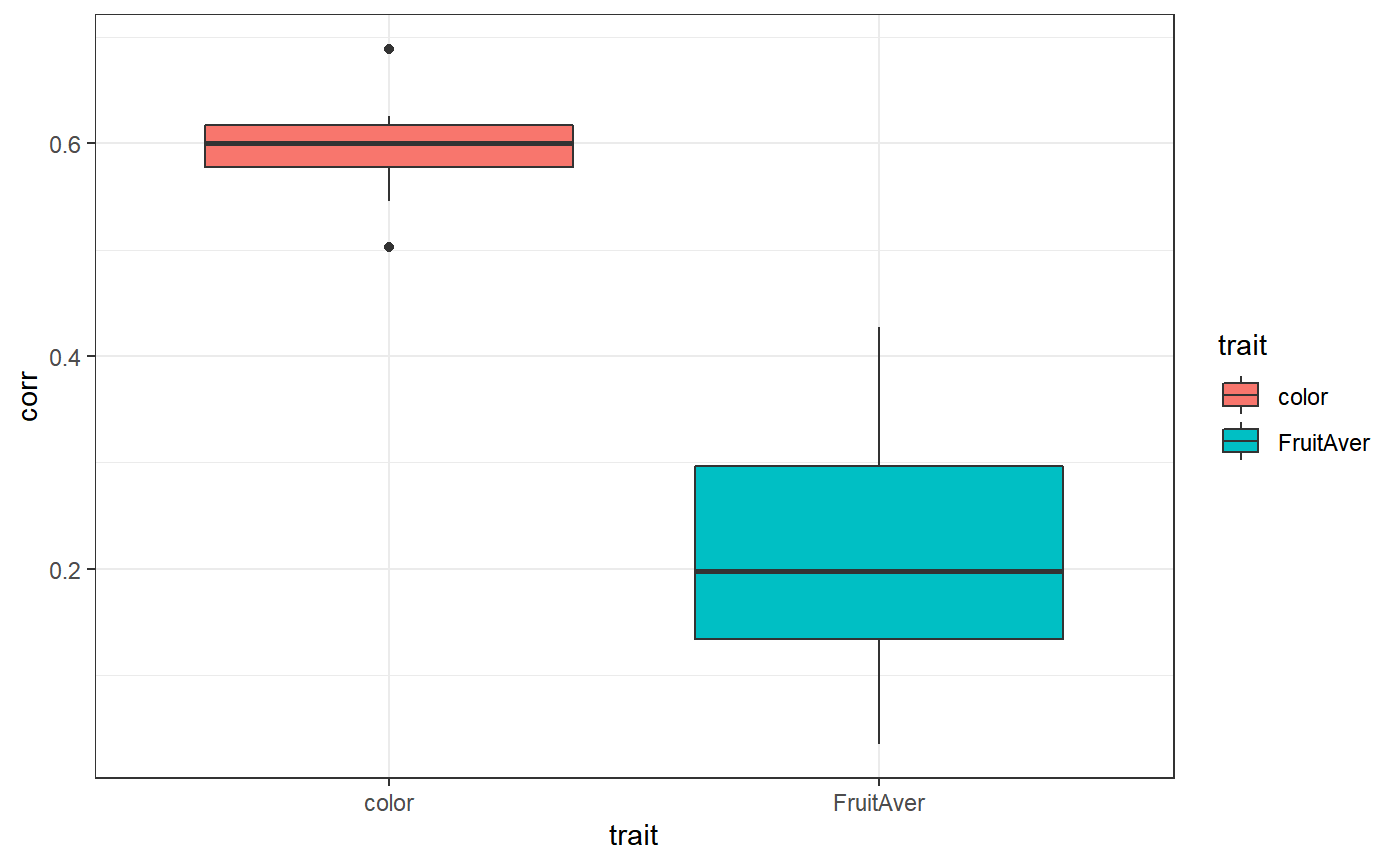
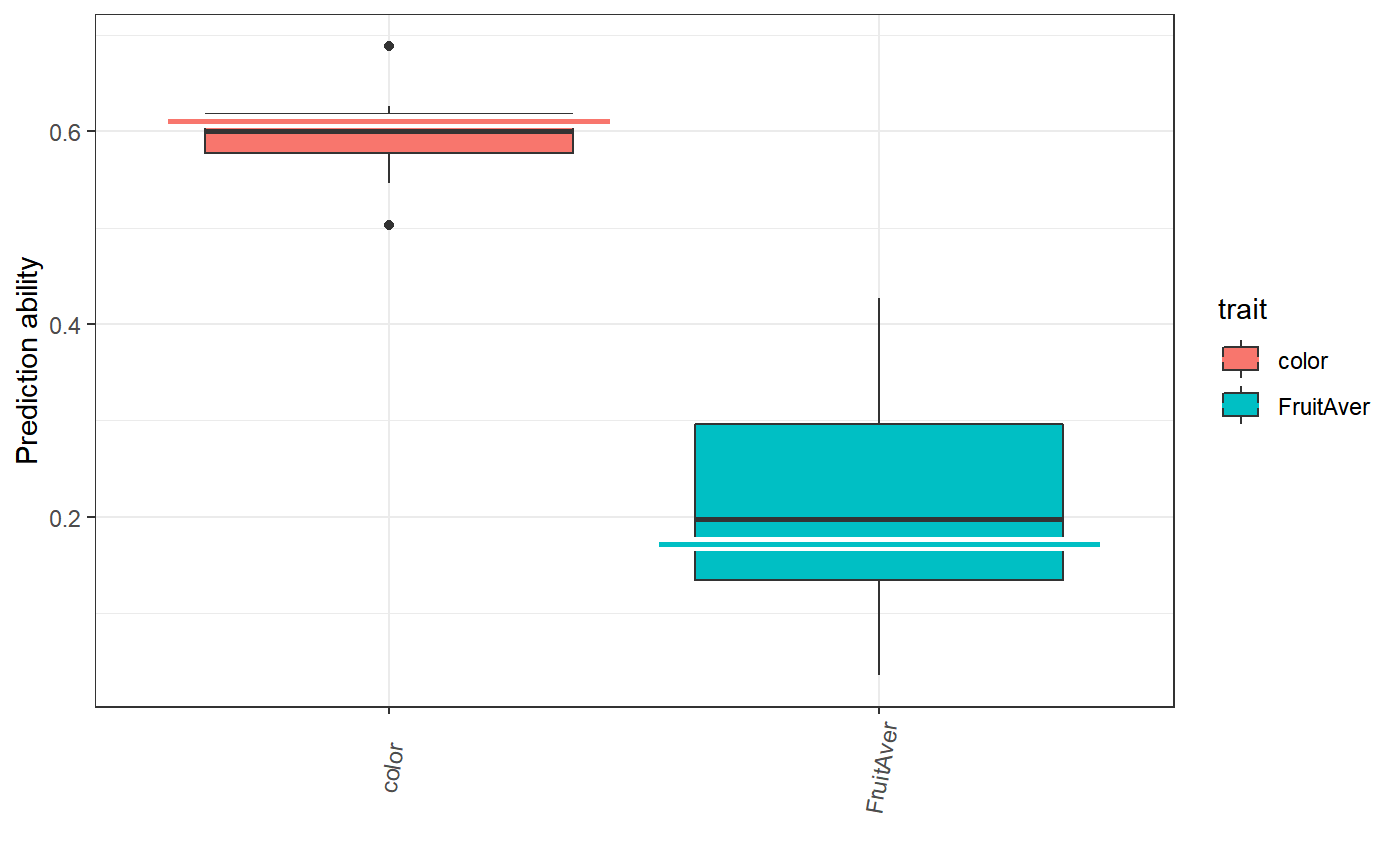Variance explained
tmp2 <- crossGP(geno,samp,phen,prior = "sommer", niter=5,testporc = 0,traits = names(phen)[c(5,7)])
# Warning: traits with zero variance were removed from phen: Year
# []==============================================================[]
# []==================== Genomic Prediction =====================[]
# []============= ASReml - BGLR - sommer package ================[]
# []======= Last update: 2020-03-18 Johan Aparicio ==============[]
# []==============================================================[]
# color == 362
# FruitAver == 362
#
# prior trait randomPop r.sqr corr finishedAt
# sommer color Pop_1 0.611 0.8777 2020-04-02 21:38:36
# sommer FruitAver Pop_1 0.1719 0.6235 2020-04-02 21:38:37
#
# []============================ End =============================[]Final plot
tmp <- tmp$data[,c("trait","corr")]
tmp2 <- tmp2$data[,c("trait","r.sqr")]
merG <- merge(tmp,tmp2, by="trait", all=T)p <- ggplot(merG, aes(x=trait,y=corr, fill=trait))+geom_boxplot()+theme_bw()+xlab(" ")+
geom_segment(data=merG,aes(x = as.numeric(as.factor(trait)) - 0.48,
xend = as.numeric(as.factor(trait)) + 0.48,
yend = r.sqr ,
y = r.sqr ),size=2.5, color="white")+
geom_segment(data=merG,aes(x = as.numeric(as.factor(trait)) - 0.45,
xend = as.numeric(as.factor(trait)) + 0.45,
yend = r.sqr ,
y = r.sqr ,
colour = trait),size=1)+
ylab("Prediction ability")+
theme(axis.text.x=element_text(angle=80,vjust=0.5))
p