Modeling plant emergence and canopy growth using UAV data
This vignette demonstrates piecewise regression using canopy data derived from UAV imagery to estimate two key parameters:
- t1: days to plant emergence.
- t2: days to reach maximum canopy.
The data are from the University of Wisconsin-Madison potato breeding program, specifically for a partially replicated experiment. The UAV images were collected in 2020 and processed in 2024.
1. Exploring data
We begin with the explorer function, which provides basic statistical summaries and descriptive statistics, as well as visualizations to help understand the temporal evolution of each plot.
names(explorer)
#> [1] "summ_vars" "summ_metadata" "locals_min_max" "dt_long"
#> [5] "metadata" "x_var"
p1 <- plot(explorer, type = "evolution", return_gg = TRUE, add_avg = TRUE)
p2 <- plot(explorer, type = "x_by_var", return_gg = TRUE)
ggarrange(p1, p2)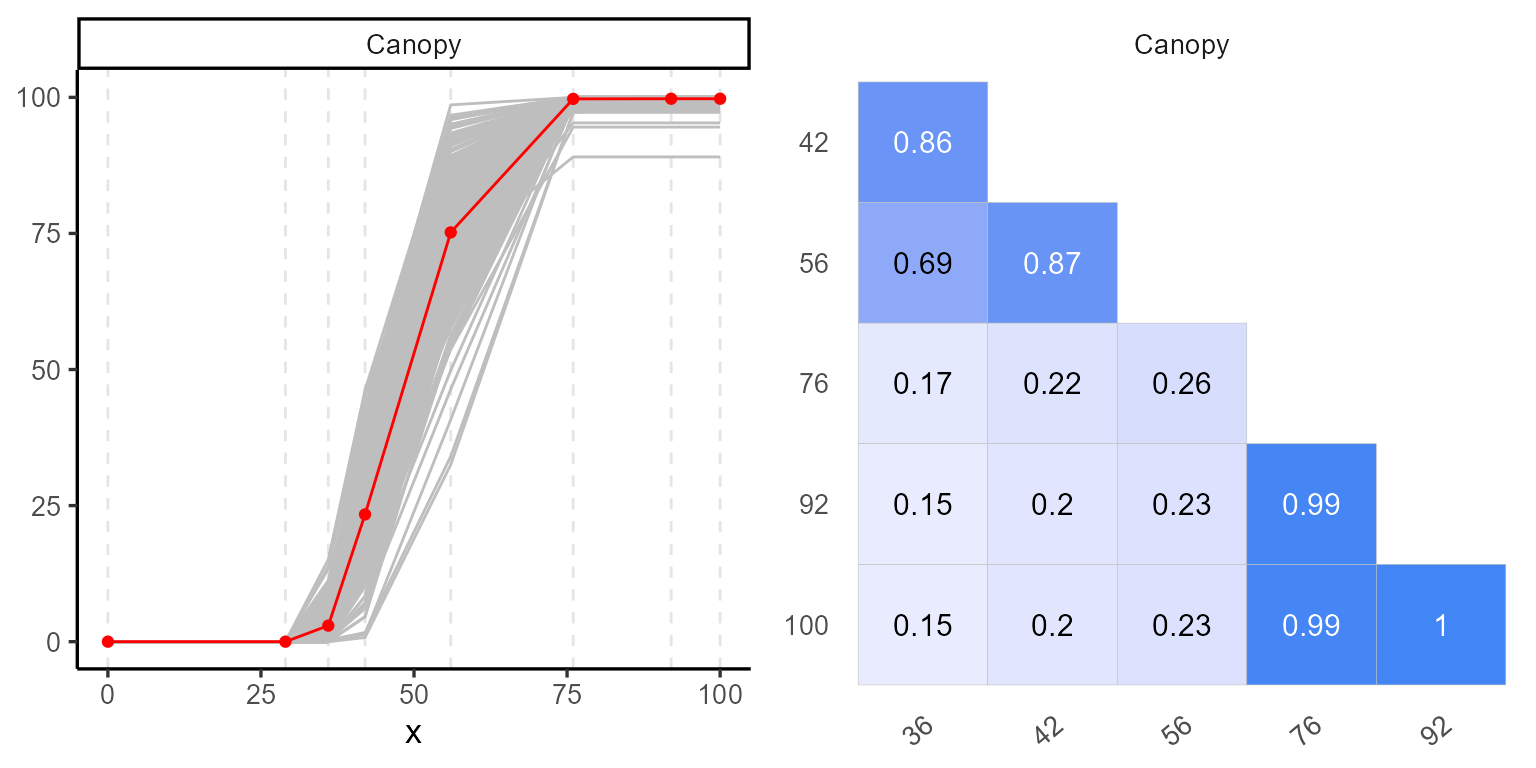
To see more about the type of plots visit
plot.explorer().
| var | x | Min | Mean | Median | Max | SD | CV | n | miss | miss% | neg% |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Canopy | 0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | NaN | 196 | 0 | 0 | 0 |
| Canopy | 29 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | NaN | 196 | 0 | 0 | 0 |
| Canopy | 36 | 0.00 | 2.95 | 1.84 | 15.09 | 3.22 | 1.09 | 196 | 0 | 0 | 0 |
| Canopy | 42 | 0.76 | 23.38 | 22.91 | 46.23 | 9.31 | 0.40 | 196 | 0 | 0 | 0 |
| Canopy | 56 | 32.51 | 75.20 | 74.96 | 98.62 | 12.26 | 0.16 | 196 | 0 | 0 | 0 |
| Canopy | 76 | 89.06 | 99.72 | 100.00 | 100.00 | 1.04 | 0.01 | 196 | 0 | 0 | 0 |
| Canopy | 92 | 89.06 | 99.75 | 100.00 | 100.04 | 1.02 | 0.01 | 196 | 0 | 0 | 0 |
| Canopy | 100 | 89.06 | 99.75 | 100.00 | 100.04 | 1.02 | 0.01 | 196 | 0 | 0 | 0 |
2. Regression Function
Once the data have been explored, we define the expectation function. In this case, it is a piece-wise regression function with three parameters: t1, t2, and k. The function can be expressed mathematically as follows:
\[\begin{equation} f(t; t_1, t_2, k) = \begin{cases} 0 & \text{if } t < t_1 \\ \dfrac{k}{t_2 - t_1} \cdot (t - t_1) & \text{if } t_1 \leq t \leq t_2 \\ k & \text{if } t > t_2 \end{cases} \end{equation}\]
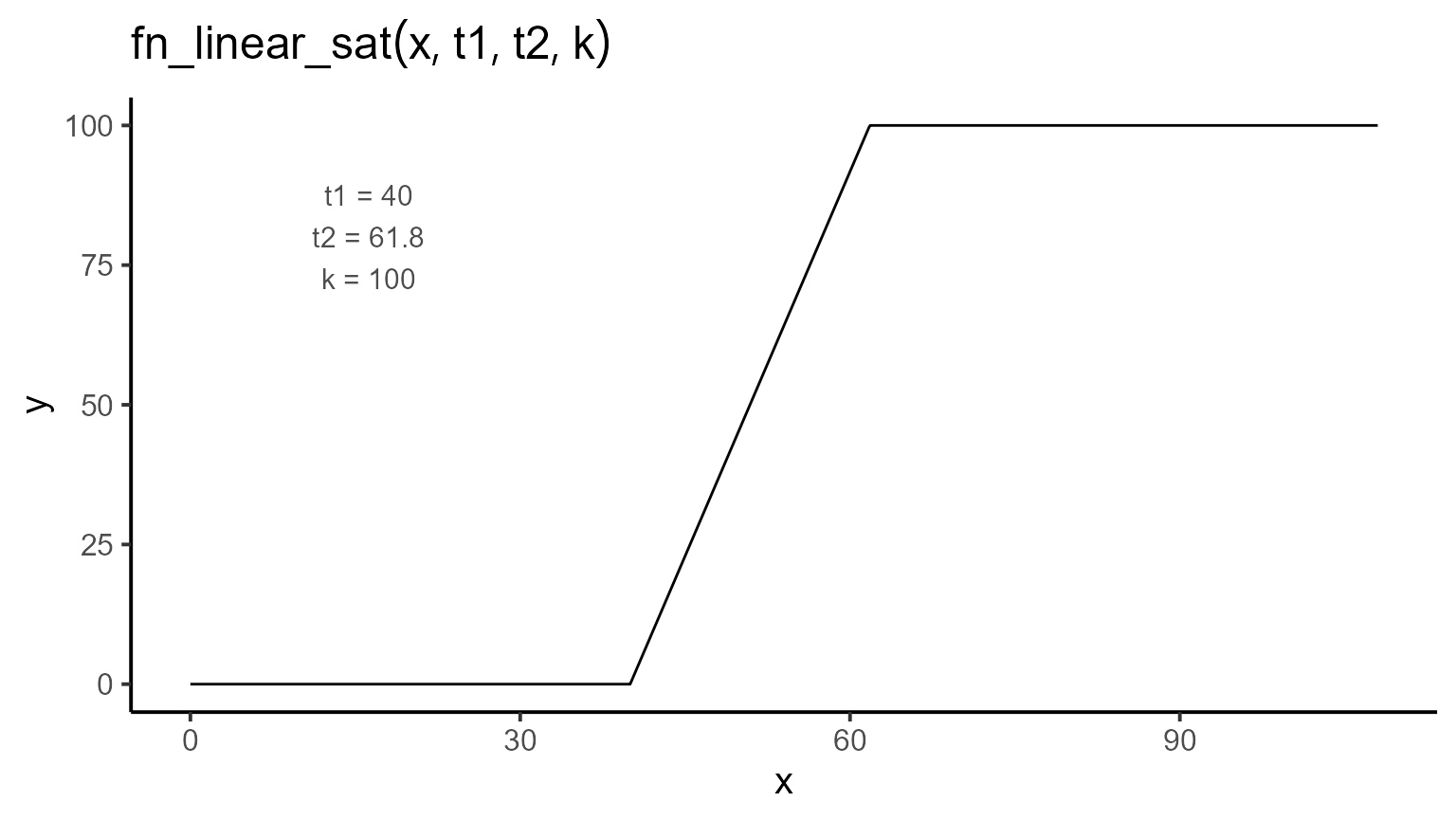
3. Fitting Models
To fit the model, we use the modeler function. Here:
- x specifies the days after planting (DAP),
- y is the canopy variable to be modeled,
- grp allows us to perform group analysis, e.g., on multiple plots.
In this example, we have 196 plots but will only fit the model for
plots 166 and 40 as a subset. We define the piecewise function
fn_lin_plat and set initial values for the parameters.
mod_1 <- dt_potato |>
modeler(
x = DAP,
y = Canopy,
grp = Plot,
fn = "fn_lin_plat",
parameters = c(t1 = 45, t2 = 80, k = 0.9),
subset = c(166, 40)
)
mod_1
#>
#> Call:
#> Canopy ~ fn_lin_plat(DAP, t1, t2, k)
#>
#> Residuals (`Standardized`):
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> -1.779e+00 -4.079e-04 1.000e-08 0.000e+00 8.157e-04 1.779e+00
#>
#> Optimization Results `head()`:
#> uid t1 t2 k sse
#> 40 34.8 60.6 100 0.0545
#> 166 31.6 57.5 100 40.9135
#>
#> Metrics:
#> Groups Timing Convergence Iterations
#> 2 0.6005 secs 100% 551.5 (id)After fitting, we can inspect the model summary and visualize the fit using the plot function:
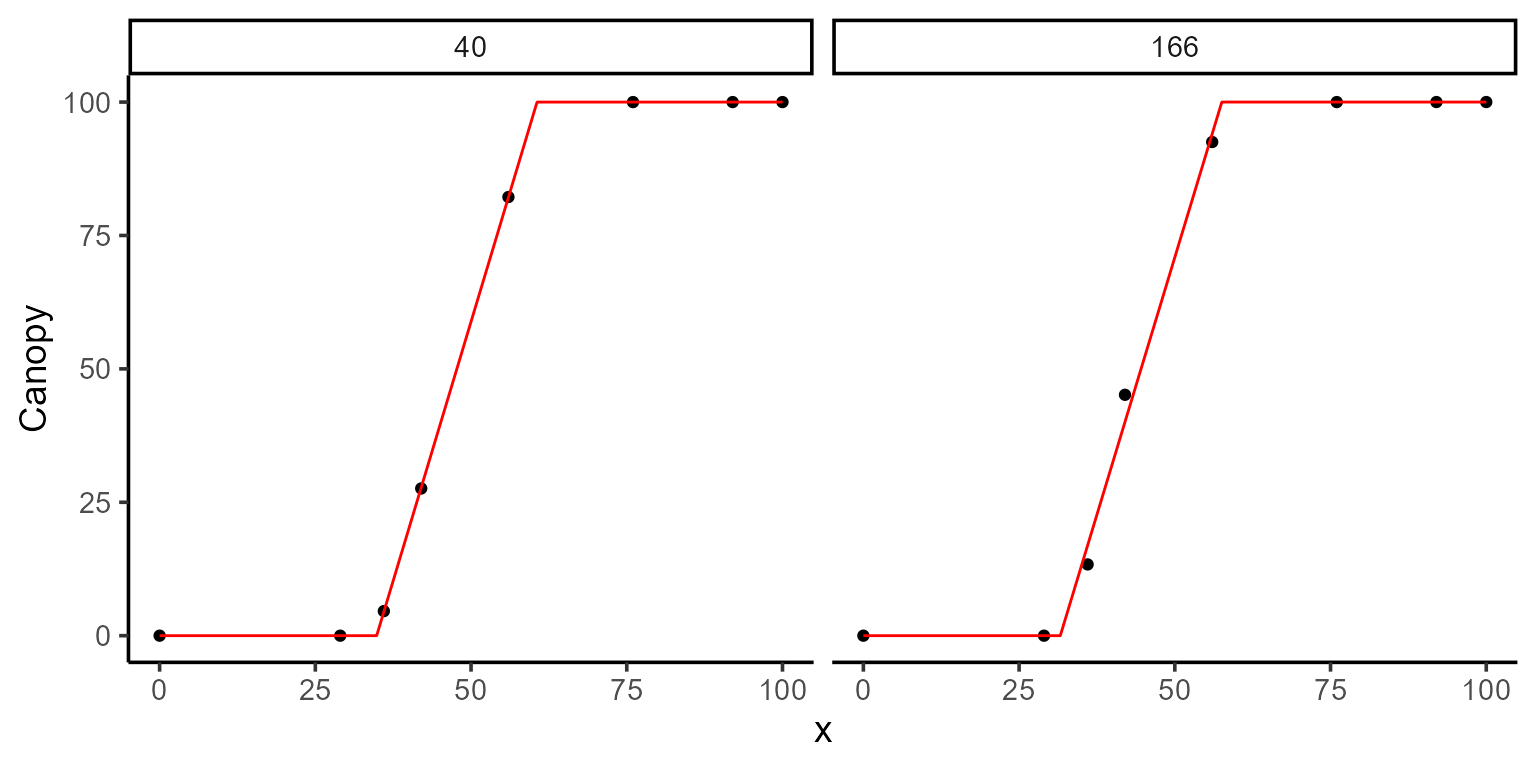
kable(mod_1$param)| uid | t1 | t2 | k | sse | fn_name |
|---|---|---|---|---|---|
| 40 | 34.84916 | 60.59505 | 100.0000 | 0.0544833 | fn_lin_plat |
| 166 | 31.61374 | 57.54603 | 100.0047 | 40.9134555 | fn_lin_plat |
3.1. Extracting model coefficients and uncertainty measures
Once the model is fitted, we can extract key statistical information, such as coefficients, standard errors, confidence intervals, and the variance-covariance matrix for each group (plot). These metrics allow us to draw conclusions about the parameter estimates and assess the uncertainty around them.
The functions coef, confint, and
vcov are used as follows:
- coef: Extracts the estimated coefficients for each group.
- confint: Provides the confidence intervals for the parameter estimates.
- vcov: Returns the variance-covariance matrix, which can be used to understand the relationships between the estimates and their variability.
coef(mod_1)
#> # A tibble: 6 × 7
#> uid fn_name coefficient solution std.error `t value` `Pr(>|t|)`
#> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 40 fn_lin_plat t1 34.8 0.0240 1453. 2.93e-15
#> 2 40 fn_lin_plat t2 60.6 0.0368 1648. 1.56e-15
#> 3 40 fn_lin_plat k 100.0 0.0603 1659. 1.51e-15
#> 4 166 fn_lin_plat t1 31.6 0.794 39.8 1.89e- 7
#> 5 166 fn_lin_plat t2 57.5 0.902 63.8 1.79e- 8
#> 6 166 fn_lin_plat k 100. 1.65 60.6 2.32e- 8
confint(mod_1)
#> # A tibble: 6 × 7
#> uid fn_name coefficient solution std.error ci_lower ci_upper
#> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 40 fn_lin_plat t1 34.8 0.0240 34.8 34.9
#> 2 40 fn_lin_plat t2 60.6 0.0368 60.5 60.7
#> 3 40 fn_lin_plat k 100.0 0.0603 99.8 100.
#> 4 166 fn_lin_plat t1 31.6 0.794 29.6 33.7
#> 5 166 fn_lin_plat t2 57.5 0.902 55.2 59.9
#> 6 166 fn_lin_plat k 100. 1.65 95.8 104.
vcov(mod_1)
#> $`40`
#> t1 t2 k
#> t1 5.755016e-04 -0.0002977975 4.429740e-08
#> t2 -2.977975e-04 0.0013525945 9.350853e-04
#> k 4.429740e-08 0.0009350853 3.632249e-03
#> attr(,"fn_name")
#> [1] "fn_lin_plat"
#>
#> $`166`
#> t1 t2 k
#> t1 0.630721491 -0.2613850 -0.002831928
#> t2 -0.261385006 0.8131631 0.711976842
#> k -0.002831928 0.7119768 2.725671099
#> attr(,"fn_name")
#> [1] "fn_lin_plat"4. Providing different initial values
The initial fit may not always be optimal, so we can adjust the initial parameter values for each plot and even fix certain parameters to improve the model.
initials <- data.frame(
uid = c(166, 40),
t1 = c(70, 60),
t2 = c(40, 80),
k = c(100, 100)
)
kable(initials)| uid | t1 | t2 | k |
|---|---|---|---|
| 166 | 70 | 40 | 100 |
| 40 | 60 | 80 | 100 |
mod_2 <- dt_potato |>
modeler(
x = DAP,
y = Canopy,
grp = Plot,
fn = "fn_lin_plat",
parameters = initials,
subset = c(166, 40)
)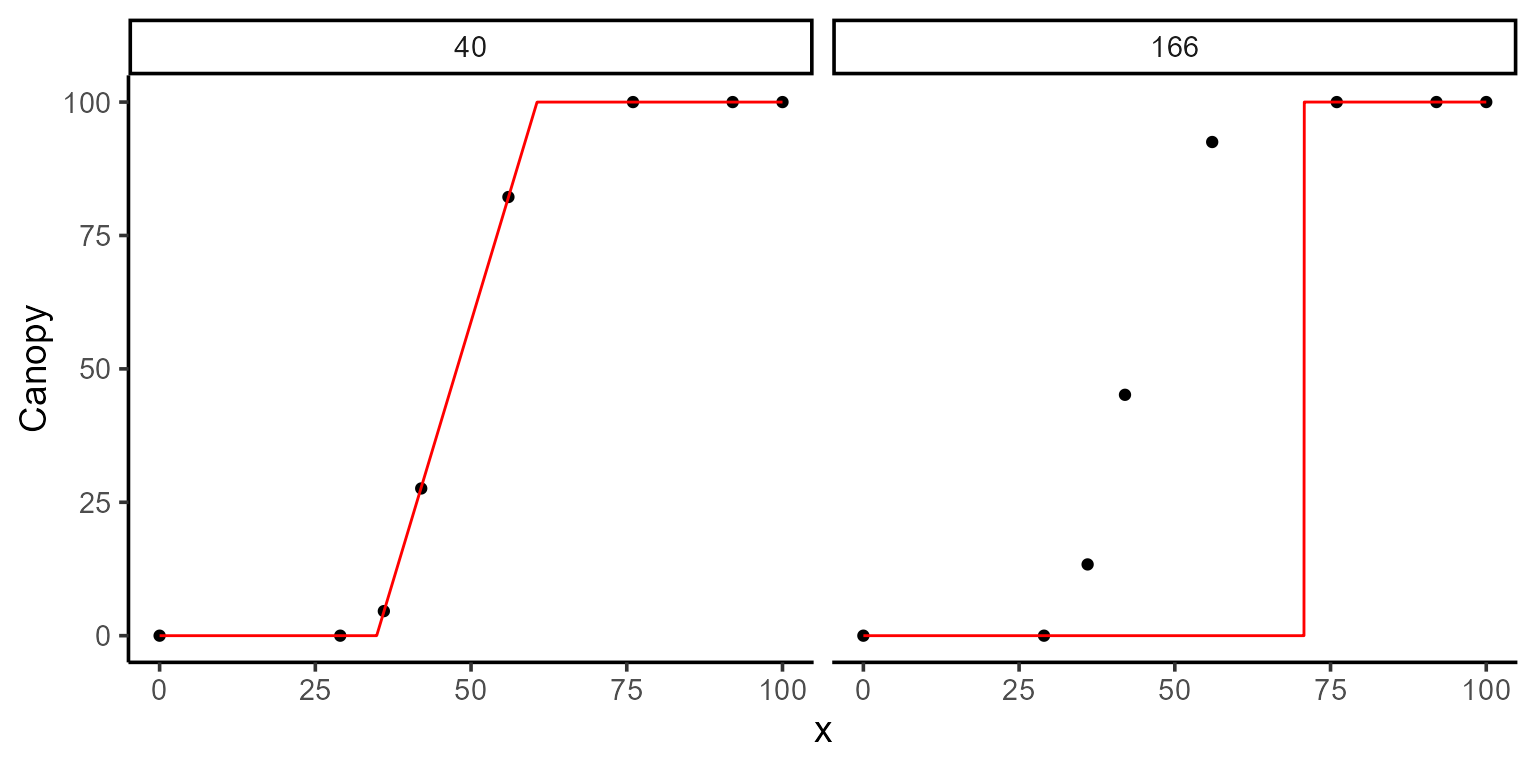
kable(mod_2$param)| uid | t1 | t2 | k | sse | fn_name |
|---|---|---|---|---|---|
| 40 | 34.84916 | 60.59505 | 100.0000 | 5.448330e-02 | fn_lin_plat |
| 166 | 70.75697 | 39.85048 | 100.0047 | 1.077531e+04 | fn_lin_plat |
It’s important to note that providing poor initial guesses for the parameters can lead to inaccurate or unreliable model fits. For example, if we mistakenly assign t1 (the day of plant emergence) a value greater than t2 (the day of maximum canopy), the model fit can fail or produce nonsensical results.
5. Fixing some parameters of the model
In certain cases, we may want to fix specific parameters either
because they are known or because we prefer the model to leave these
parameters unchanged. For example, we can fix the parameter
k, which represents the maximum canopy value, as
follows:
fixed_params <- list(k = "max(y)")
mod_3 <- dt_potato |>
modeler(
x = DAP,
y = Canopy,
grp = Plot,
fn = "fn_lin_plat",
parameters = c(t1 = 45, t2 = 80, k = 0.9),
fixed_params = fixed_params,
subset = c(166, 40)
)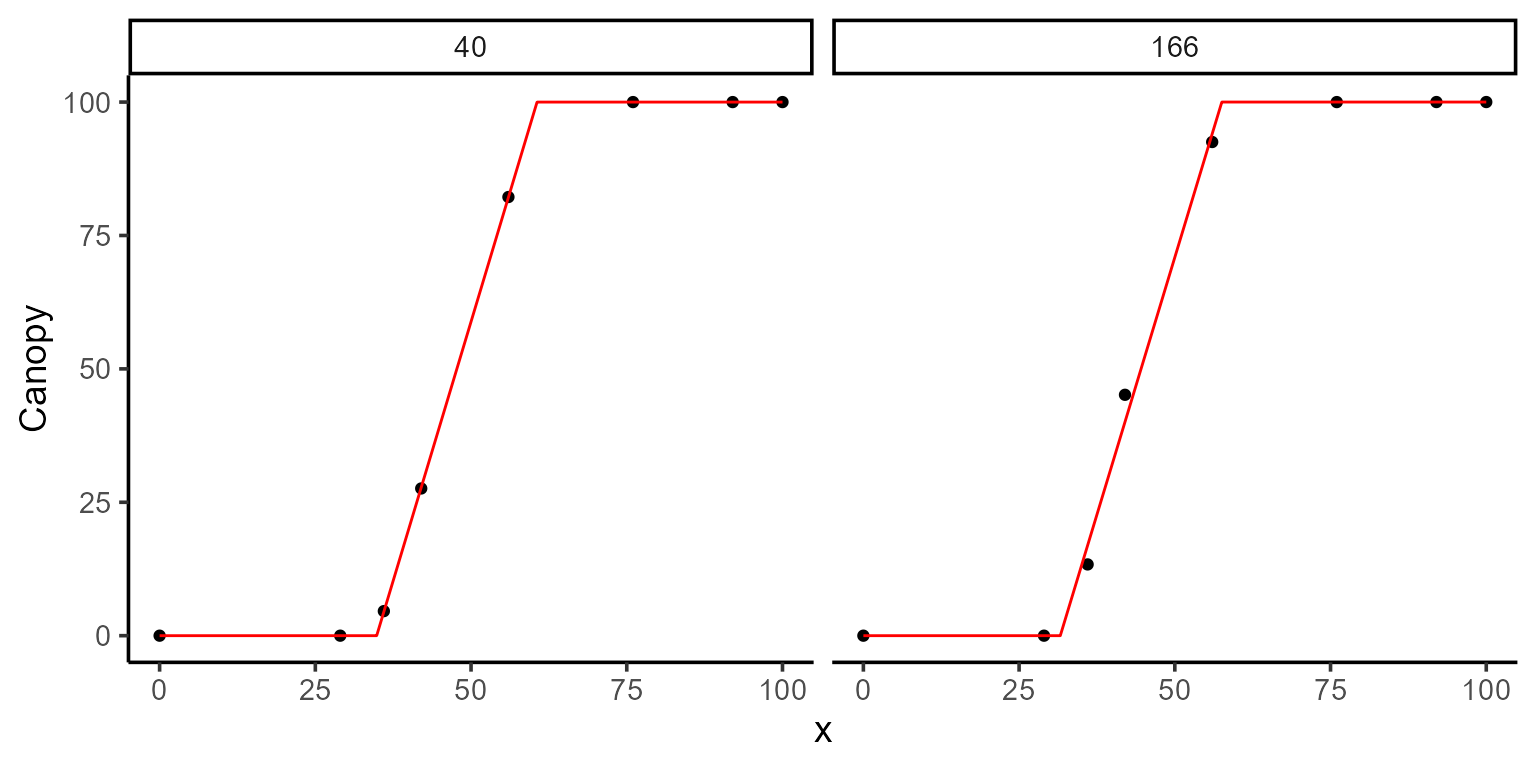
kable(mod_3$param)| uid | t1 | t2 | sse | k | fn_name |
|---|---|---|---|---|---|
| 40 | 34.84916 | 60.59505 | 0.0544833 | 100.000 | fn_lin_plat |
| 166 | 31.61374 | 57.54663 | 40.9134718 | 100.007 | fn_lin_plat |
By fixing k to 100, we are telling the model that the maximum canopy for these plots is fixed at 100%. This allows the model to focus on estimating the other parameters, t1 and t2, potentially improving the accuracy of their estimates by reducing the complexity of the model.
6. Comparing estimations
rbind.data.frame(
mutate(mod_1$param, model = "1", .before = uid),
mutate(mod_2$param, model = "2", .before = uid),
mutate(mod_3$param, model = "3", .before = uid)
) |>
filter(uid %in% 166) |>
kable()| model | uid | t1 | t2 | k | sse | fn_name |
|---|---|---|---|---|---|---|
| 1 | 166 | 31.61374 | 57.54603 | 100.0047 | 40.91346 | fn_lin_plat |
| 2 | 166 | 70.75697 | 39.85048 | 100.0047 | 10775.31306 | fn_lin_plat |
| 3 | 166 | 31.61374 | 57.54663 | 100.0070 | 40.91347 | fn_lin_plat |
After fitting multiple models with different initial values, fixed
parameters, and canopy adjustments, we can compare the resulting
coefficients and sum of square errors (sse) to evaluate the
impact of these changes.
comparison <- performance(mod_1, mod_2, mod_3)
comparison |>
filter(uid %in% 166) |>
kable()| fn_name | uid | df | nobs | p | logLik | AIC | AICc | BIC | Sigma | SSE | MAE | MSE | RMSE | R2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fn_lin_plat_1 | 166 | 4 | 8 | 3 | -17.88 | 43.76 | 57.09 | 44.08 | 2.86 | 40.91 | 1.27 | 5.11 | 2.26 | 1.0 |
| fn_lin_plat_2 | 166 | 4 | 8 | 3 | -40.17 | 88.35 | 101.68 | 88.67 | 46.42 | 10775.31 | 18.88 | 1346.91 | 36.70 | 0.3 |
| fn_lin_plat_3 | 166 | 3 | 8 | 2 | -17.88 | 41.76 | 47.76 | 42.00 | 2.61 | 40.91 | 1.27 | 5.11 | 2.26 | 1.0 |
plot(comparison, id = 166)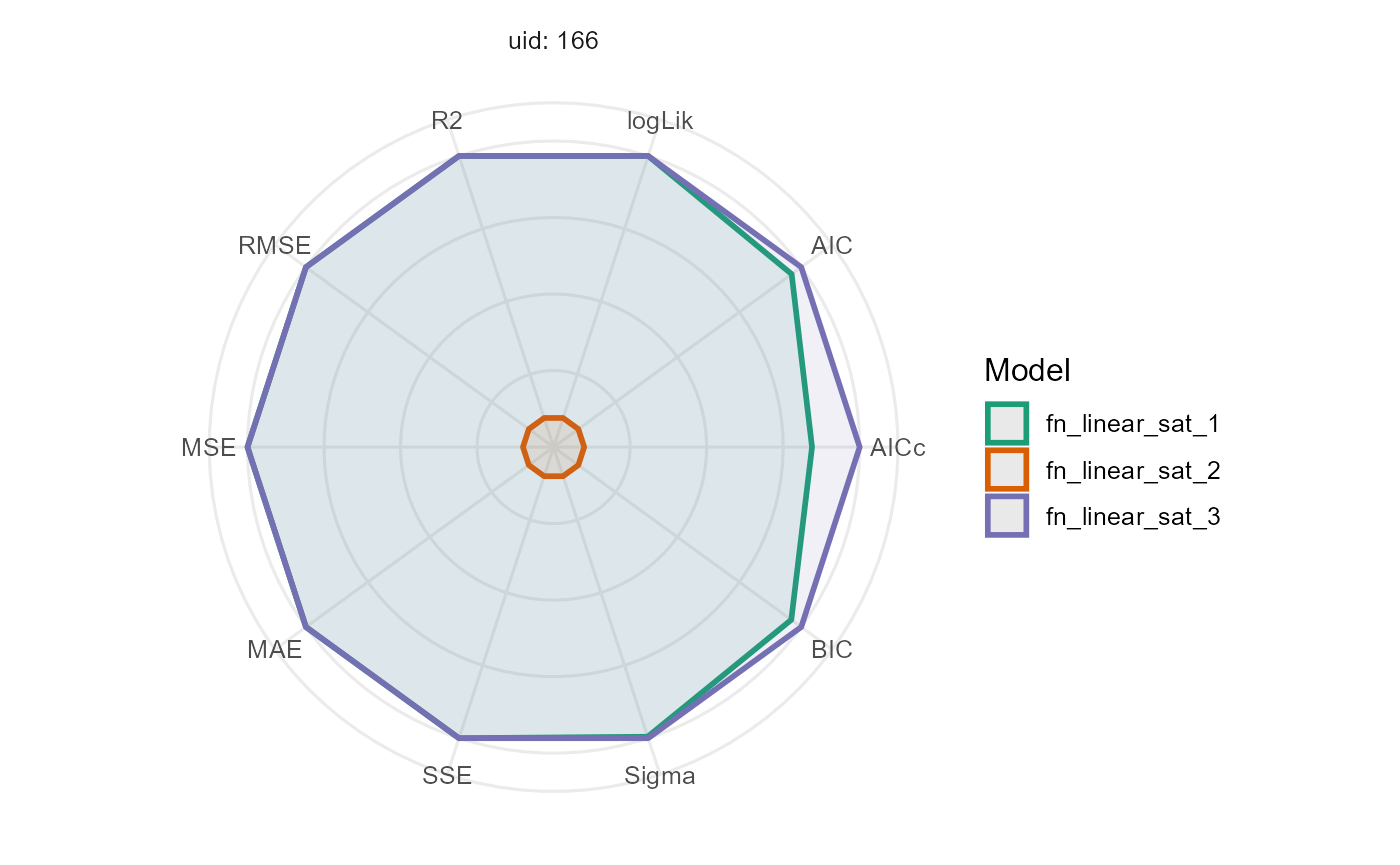
7. Making predictions
Once the model is fitted and validated as the best representation of
our data, we can proceed to make predictions. The
predict.modeler() function provides a range of flexible
prediction options, allowing users to perform point predictions,
calculate the area under the curve (AUC), compute first or second
derivatives, and even evaluate custom functions of the parameters. Below
are some examples demonstrating these capabilities:
| uid | fn_name | x_new | predicted.value | std.error |
|---|---|---|---|---|
| 166 | fn_lin_plat | 45 | 51.62246 | 1.656734 |
| uid | fn_name | x_min | x_max | predicted.value | std.error |
|---|---|---|---|---|---|
| 166 | fn_lin_plat | 0 | 108 | 6342.308 | 93.61781 |
| uid | fn_name | formula | predicted.value | std.error |
|---|---|---|---|---|
| 166 | fn_lin_plat | t2 - t1 | 25.93229 | 1.402375 |
In each example, the predict.modeler() function tailors
the predictions to the user’s needs, whether it’s estimating a single
value, integrating across a range, or calculating a parameter-based
expression.
8. Modelling all plots using parallel processing
Finally, we can apply this method to all 196 plots, leveraging
parallel processing to speed up the computation. To do this, we specify
parallel = TRUE in the options argument, and set the number
of cores using the function parallel::detectCores(), which
automatically detects the available cores.
mod <- dt_potato |>
modeler(
x = DAP,
y = Canopy,
grp = Plot,
fn = "fn_lin_plat",
parameters = c(t1 = 45, t2 = 80, k = 0.9),
fixed_params = list(k = "max(y)"),
options = list(progress = TRUE, parallel = TRUE, workers = 5)
)